2. 中国科学院微生物研究所, 北京 100101
2. Institute of Microbiology, Chinese Academy of Sciences, Beijing, 100101, China
染色体携带了生物体的遗传信息,并在细胞中代代相传,保证了各个物种的遗传稳定性。在细胞的分裂过程中染色体必须被正确地拷贝和精确地分配,同时修复存在的DNA损伤,确保新的细胞中含有与亲代细胞相同的遗传信息。在人体的1×1014个细胞中每天都发生数千个DNA损伤,这些损伤可能源于代谢副产品活性氧族引起的损伤或DNA复制过程中的失误,也可能源于紫外线(UV)照射、电离辐射(Ionizing Radiation, IR)或化学物质等外界因素[1]。这些DNA损伤都有导致基因突变的潜在可能,而未修复的基因突变可能通过RNA和蛋白质表达影响细胞的功能,并遗传给子代细胞,引起细胞衰老、凋亡和癌变等。DNA损伤修复机制已成为遗传学领域研究的热点。近年来的研究发现,染色体结构维持蛋白(Structure Maintenance of Chromosome, Smc)在维持染色体结构及DNA损伤修复方面发挥关键作用。Smc蛋白家族包括3类:黏连蛋白(Cohesin),凝缩蛋白(Condesin)和Smc5/6复合体(Smc5/6 complex)。黏连蛋白主要在细胞分裂前期发挥作用,将新复制的两条姐妹染色单体聚集在一起,促进姐妹染色单体的重组;凝缩蛋白主要参与细胞分裂中期染色体的凝缩过程;Smc5/6复合体在细胞周期的调控、有丝分裂和减数分裂的DNA损伤修复、维持基因组的稳定性方面都具有重要的调控作用[2-4]。目前对黏连蛋白和凝缩蛋白的功能研究比较深入,而对Smc5/6复合体的功能和分子机理研究相对较少。因此,深入研究Smc5/6复合体的结构和各亚基的功能,对进一步理解DNA损伤修复机制及基因组稳定性的分子机制具有重要的指导意义。本文综述了近年来对Smc5/6复合体的研究进展,从Smc5/6复合体的结构特征和生物学功能两个方面进行阐述。
1 Smc5/6复合体的结构特征 1.1 Smc5/6复合体的组件蛋白Smc5/6复合体首次被发现是在裂殖酵母中鉴定到的具有Smc蛋白结构的Rad18和Spr18的异质二聚体[5]。Smc5/6蛋白复合体由两个Smc蛋白(Smc5和Smc6)异二聚体及非Smc蛋白亚基共同组成,其中在酵母、拟南芥和人体细胞中已有6个非Smc亚基(Nse1-6)被鉴定出来[6-9]。Smc蛋白是由1 000-1 300个氨基酸残基组成的多肽,在N-端和C-端分别有一个球状的结构域,而在Smc蛋白中间含有一个铰链(Hinge)结构域。N-端和C-端的球状结构域由中间的铰链结构域以反向平行的相互作用方式聚集在一起,产生一个具有ATP酶活性的球状头部(Heads)结构域[10, 11]。Smc5和Smc6蛋白在不同物种之间相对保守,具有相似的Smc蛋白结构特征。由于Nse1的C-端含有一个Ring-like结构域,因此推测Nse1可能具有泛素化蛋白酶的特性。研究发现Nse1单独作用时并不具有泛素化蛋白酶活性,但有Nse3同时存在时表现出微弱的泛素化蛋白酶活性[12]。Nse2/Mms21最初在出芽酵母中通过遗传筛选鉴定得到[13],Nse2/Mms21的C-端也包含一个Ring结构域,赋予了Nse2/Mms21的SUMO连接酶活性[14, 15]。Nse3的C端含有一个mage功能域,研究发现该结构域具有DNA结合特性,nse3基因在真菌及植物中是单拷贝,但是在人体细胞中与具有多拷贝的黑色素瘤抗原基因(mage)有很高的相似度,研究发现人体细胞中只有Mage-G1蛋白能与Smc5/6复合体发生免疫共沉淀[16-18]。Nse4是具有螺旋翼状域的结构蛋白,是Smc5/6复合体的必需蛋白,作为桥梁结合到Smc蛋白上稳定复合体的结构,在维持整个复合体的稳定性方面发挥关键作用[19]。Nse5和Nse6蛋白是Smc5/6复合体的非保守部分,目前仅在酵母、拟南芥和人体细胞鉴定到。
1.2 Smc5/6复合体的结构特征整个Smc5/6蛋白复合体由3个亚复合体(Smc5-Smc6-Nse2、Nse1-Nse3-Nse4、Nse5-Nse6)按照特定的形式组合而成[19-21](图 1)。Smc5-Smc6-Nse2亚复合体是整个Smc5/6复合体的核心组件,构成了整个复合体的支架。Smc5和Smc6两个蛋白通过其铰链结构域相互作用形成异二聚体(Smc5-Smc6),Nse2/Mms21通过其N-末端与Smc5的卷曲螺旋结构域结合形成Smc5-Smc6-Nse2亚复合体[14, 22]。Nse1、Nse3和Nse4形成的Nse1-Nse3-Nse4亚复合体具有高度的保守性,结合在Smc5和Smc6的球状头部结构域和DNA分子,该三聚体的稳定性对于Smc5/6复合体在指定的DNA损伤位点聚集是必需的。Nse1通过Ring-like结构域与Nse3的N-端相互作用增加其对双链DNA的结合作用[23]。Nse3的C-端与Nse4直接相互作用进一步形成异源三聚体Nse1-Nse3-Nse4[24]。Nse4一方面与Nse3结合形成Nse1-Nse3-Nse4异源三聚体,另一方面与Smc5和Smc6的头部相结合使得Smc5/6复合体形成一个封闭的环状结构[19]。Nse5-Nse6二聚体在不同的生物体中具有较大差异。Nse5和Nse6在裂殖酵母中以二聚体的形式结合在Smc5/6复合物的球形头部区域附近,在芽殖酵母中Nse5和Nse6与Smc5/6的铰链区相互作用,而在拟南芥和人体细胞中这两个蛋白的定位还未知[8, 20, 25]。近期Taschner等[26]研究发现,在芽殖酵母中Nse5-Nse6二聚体具有头部和铰链区两个结合位点,通过抑制Smc5/6复合体的ATP水解酶活性,对Smc5/6复合体结合DNA底物具有重要的调控作用。
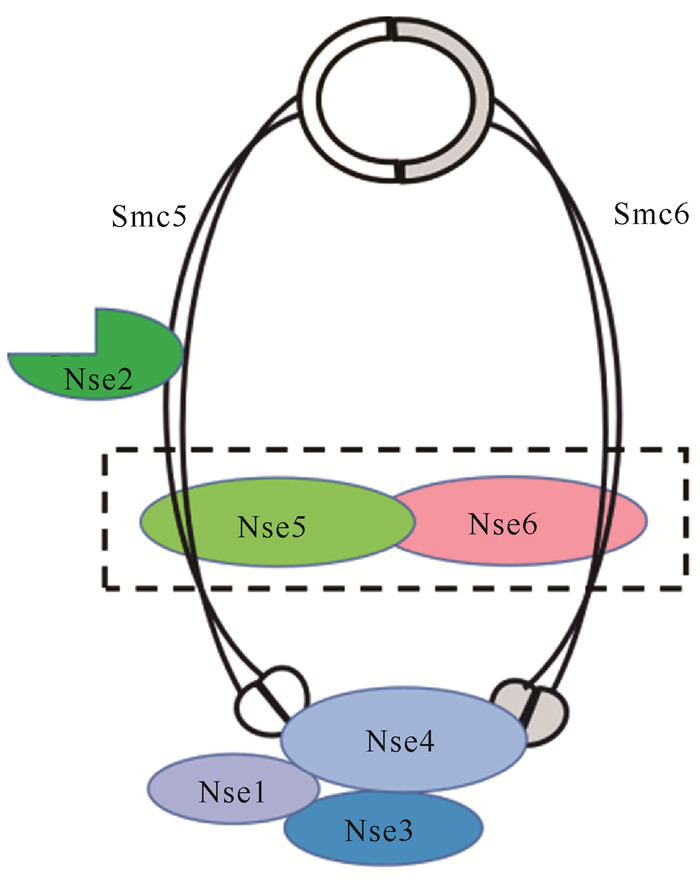
|
| Smc5/6由Smc5、Smc6和非Smc亚基构成,其中Nse1、Nse2 (Mms21)、Nse3 (Mage-G1)和Nse4 4个亚基在真核生物中高度保守,而Nse5和Nse6在序列上不保守,目前仅在酵母、拟南芥和人体细胞中被发现 The Smc5/6 complex consists of Smc5, Smc6 and the non-Smc elements (NSE), among which Nse1, Nse2 (Mms21), Nse3 (Mage-G1), and Nse4 are highly conserved across eukaryotes, whereas Nse5 and Nse6 are not known to be conserved at the level of sequence, as they have been identified only in yeast, Arabidopsis and human cells 图 1 Smc5/6复合体的结构 Fig. 1 Structure of Smc5/6 complex |
2 Smc5/6复合体的生物学功能研究
对Smc5/6复合体的生物学功能研究相对较少,近年来在真菌、动物和植物等模式生物中的研究发现,Smc5/6复合体在DNA损伤修复、缓解复制压力、维持端粒长度、维持细胞的正常分裂和分化等方面具有重要作用[27]。
2.1 参与DNA损伤修复细胞由于外界环境或自身因素的影响会导致各种DNA损伤,其中DNA双链断裂(Double Stand Break, DSB)是一种严重的DNA损伤,机体中未能修复的DSB将严重影响个体的生长和发育。在细胞中主要存在两种DSB修复途径:非同源重组末端连接(Non-Homologous End Joining, NHEJ)和同源重组修复(Homologous Recombination, HR)。NHEJ主要在细胞有丝分裂G1期发挥作用,直接对断裂部位通过末端连接的方式完成DSB的修复。HR主要发生在细胞有丝分裂G2/S期,利用染色体为模板以同源重组的方式完成DSB的修复,这是细胞中最精确的DNA修复途径。近年来研究发现,Smc5/6复合体在DNA同源重组修复方面发挥重要作用,在裂殖酵母S.pombe中nse1基因缺失导致对由电离辐射(IR)、紫外线(UV)和甲基磺酸甲酯(MMS)引起的DNA损伤修复缺陷[12, 28],裂殖酵母中Smc5/6能与同源重组关键蛋白Rad51/Rhp51相互作用,表明Smc5/6在同源重组修复中具有重要的作用[8, 29]。在酿酒酵母S.cerevisiae有丝分裂G2期,Smc5/6通过影响Rad50/Mre11聚集在DSB损伤位点来影响姐妹染色单体重组完成DSB的修复[30, 31]。在出芽酵母中,Smc5/6蛋白复合体能直接与RecQ解螺旋酶Sgs1结合,进而引导Sgs1定位于DNA损伤位点,形成STR复合体(Sgs1-Top3-Rmi1),该复合体在DSB同源重组修复中起着关键的调控作用[32, 33]。进一步研究发现,Smc5/6复合体不仅在真菌中,而且在动物和植物细胞中也参与DSB的同源重组修复[30, 34-37]。在人体细胞中,Nse2通过Smc6和DNA修复蛋白TRAX的类泛素化促进DNA同源重组修复功能[14]。此外,Nse2通过介导Smc5蛋白的泛素化作用,在DNA损伤修复、DNA复制和维持基因组稳定性方面发挥重要作用[38, 39]。在非洲爪蟾卵细胞中鉴定得到两个新的蛋白因子Slf1和Slf2,与Rad18蛋白形成Rad18-Slf1-Slf-2三聚体复合物,招募Smc5/6复合体到DNA损伤位点降低基因组的不稳定性[40]。在拟南芥中,Nse1、Nse3和Nse4蛋白功能的缺失导致严重的DNA损伤修复缺陷[41-43]。虽然在人体和酵母细胞中Nse1没有检测到泛素化酶活性,但Nse1对于DNA损伤修复、染色体复制及稳定性是必需的[12]。
2.2 缓解DNA复制压力细胞内DNA复制不仅受到细胞代谢产物的影响,而且由于DNA复制过程中超螺旋结构的积累和姐妹染色体缠绕(SCIs)形成的拓扑压力会导致DNA复制无法顺利进行,只有去除DNA超螺旋结构和染色体缠绕才能使得DNA复制顺利进行[44]。研究发现Smc5/6复合体在去除DNA超螺旋结构的积累及姐妹染色体缠绕形成的拓扑压力过程中发挥重要作用,通过去除复制过程产生的有害中间体或者重新启动受阻的复制叉进而保证DNA复制顺利进行[27, 45]。通过对Smc5/6复合体的定位和表型分析,发现Smc5/6复合体参与DNA的复制过程:在酿酒酵母有丝分裂S期,Smc5/6存在于染色体的复制初始位点,而在G2/M期其分布方式类似于黏连蛋白的分布方式,Smc5/6存在方式的改变表明其与复制叉有关系并跟随复制叉一起前行[31, 46]。酿酒酵母中拓扑异构酶Top1和Top2协同作用去除DNA的超螺旋结构及SCIs[46],而在人体细胞中Smc5/6复合体功能的缺失导致拓扑异构酶Top2异常分布,从而进一步导致超螺旋结构、SCIs积累及染色体的异常分离[47], 这一结果表明Smc5/6复合体通过影响拓扑异构酶的分布进一步参与DNA的超螺旋结构及SCIs的去除。此外,Smc5/6复合体在降解由DNA复制引起的有害重组中间体以及重新启动复制叉的前行中也具有重要作用[48]。在DNA复制的模板置换过程中,DNA的模板链和合成链发生相互交连形成“X”型的HJ中间体,从而导致DNA复制无法正常进行。酿酒酵母中Smc5/6复合体定位分析发现其存在于终止的复制叉位点,且在Smc5/6突变体中发现“X”型重组中间体的积累,表明Smc5/6复合体可能与重组中间体的修复相关[32]。进一步研究发现,酵母中RecQ型解旋酶(Sgs1)通过与多聚SUMO链和Smc5/6复合体的SUMO E3复合体相互作用,利用泛素化途径降解同源重组形成的中间体[32, 33]。近年来研究发现,Nse5和Nse6蛋白在减数分裂的DNA重组中间体的清除和缓解复制压力方面具有重要作用[49, 50]。在裂殖酵母中Smc5/6复合体的SUMO连接酶活性及其与染色体的结合依赖于Nse5-Nse6二聚体,而且Brc1与Nse5-Nse6二聚体发生相互作用,引导Smc5/6复合体定位于DNA复制叉位点,促进Smc5/6复合体对DNA复制过程产生的错误复制叉的清除[51]。
2.3 维持端粒长度Smc5/6复合体在维持端粒长度方面的研究相对较少,只在酵母细胞及人体细胞中有少量报道[52-54]。研究发现Smc5/6复合体存在于染色体的末端[55]。在酵母细胞中,组成Smc5/6复合体的Nse2、Smc5和Smc6蛋白功能的缺失会导致端粒的缩短以及衰老[52]。在端粒酶阳性细胞中,smc6-9基因缺失在染色体终端表现出由于同源重组缺陷引起的重复序列错误分离。另外,mms21-11基因缺失表现出端粒聚集缺陷进而增加端粒位置效应(TPE)[15, 52, 53]。在端粒酶阴性细胞中,smc6-9和mms21-11等位基因突变加速了由于重组中间体积累引起的细胞衰老及DNA复制提前终止[55]。Sir4是端粒位置效应的一个重要调节因子,研究发现Smc5/6复合体能与Sir4发生相互作用,对端粒位置效应有重要影响[56]。
2.4 维持细胞正常分裂细胞的正常分裂是生物体生长发育的基础,研究发现Smc5/6复合体对细胞的正常分裂具有重要作用。在模式植物拟南芥中,Nse2通过对细胞周期及细胞分裂素信号通道的调控来维持根部细胞的正常分裂[57]。Nse1和Nse3蛋白功能的缺失导致严重的细胞分裂异常,进而影响植株胚胎的发育及种子的形成[41]。Nse3在有丝分裂染色体的正常分离过程中也发挥重要作用[7]。在模式植物拟南芥中发现两个功能保守的Nse4同源蛋白(Nse4A和Nse4B),nse4突变株在减数分裂染色体分离、植株生长和种子形成方面存在严重缺陷,表明Nse4蛋白在维持正常的减数分裂、植株的生长和种子形成过程中具有重要作用[42, 43]。在拟南芥植株中同时敲除smc6a和smc6b基因将严重影响植株配子和种子的形成[58]。通过拟南芥自然变异植株,鉴定得到Smc5/6复合体亚蛋白Sni1,其对细胞减数分裂交叉点分布具有调控作用,揭示Smc5/6复合体在植物减数分裂同源重组中具有重要的功能[59]。
2.5 其他功能近期研究表明,Smc5/6复合体不仅在DNA损伤修复、DNA复制、维持端粒长度和正常细胞分裂等方面发挥重要作用,而且在维持核糖体功能、细胞凋亡调控及抑制病毒增殖等方面也发挥重要作用。在酿酒酵母中,Nse2(Mms21)功能的缺失导致核糖体RNA产量减少和核糖体蛋白在细胞核中的积累,表明Nse2对维持细胞核的正常功能也具有重要作用[60]。近期在玉米中对Nse2同源蛋白(MMS21)的SUMO化进行了研究,发现其通过影响染色质结构进而影响基因的转录调控,导致种子致死、花粉和种子萌发严重受损[61]。在网柄菌Dictyostelium discoideum中,通过RNA干扰降低Nse4蛋白的表达,增加了紫外照射引起的细胞凋亡和由饥饿诱导形成的子实体的数目[62]。此外,Smc5/6复合体作为病毒抑制子在控制病毒增殖方面具有重要作用[63, 64]。
3 总结本文从Smc5/6复合体的结构特征和生物学功能两个方面对近年来Smc5/6复合体的研究进展进行了阐述。Smc5/6复合体在DSB同源重组修复、消除DNA复制压力、去除有害的复制中间体及维持端粒的长度等方面发挥重要作用。此外,Smc5/6复合体在胚胎发育、维持核糖体功能和抑制病毒增殖等方面也有相关报道,但其作用机制尚不清楚[34]。关于Smc5/6复合体的结构和功能在酵母细胞中的报道相对较多,在植物中的报道较少,而在整体动物模型水平上几乎没有报道。对Smc5/6各亚基的功能研究较少,特别是Nse1、Nse3、Nse4、Nse5、Nse6蛋白亚基的分子作用机制在很大程度上是未知的。因此,关于Smc5/6复合体和各亚基在植物和整体动物模型中的功能与分子作用机理还有待进一步探索。
| [1] |
罗瑛, 宋宜, 林德玲. 基因转录后调控在DNA损伤反应中的重要功能[J]. 遗传, 2014, 36(4): 309-315. |
| [2] |
WU N, YU H T. The Smc complexes in DNA damage response[J]. Cell & Bioscience, 2012, 2(1): 5. DOI:10.1186/2045-3701-2-5 |
| [3] |
LOSADA A, HIRANO T. Dynamic molecular linkers of the genome: The first decade of SMC proteins[J]. Genes < Development, 2005, 19(11): 1269-1287. |
| [4] |
JEPPSSON K, KANNO T, SHIRAHIGE K, et al. The maintenance of chromosome structure: Positioning and functioning of SMC complexes[J]. Nature Reviews Molecular Cell Biology, 2014, 15(9): 601-614. DOI:10.1038/nrm3857 |
| [5] |
LEHMANN A R, WALICKA M, GRIFFITHS D J, et al. The Rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair[J]. Molecular and Cellular Biology, 1995, 15(12): 7067-80. DOI:10.1128/MCB.15.12.7067 |
| [6] |
FOUSTERI M I, LEHMANN A R. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein[J]. The EMBO Journal, 2000, 19(7): 1691-702. DOI:10.1093/emboj/19.7.1691 |
| [7] |
PEBERNARD S, MCDONALD W H, PAVLOVA Y, et al. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis[J]. Molecular Biology of the Cell, 2004, 15(11): 4866-76. DOI:10.1091/mbc.e04-05-0436 |
| [8] |
PEBERNARD S, WOHLSCHLEGEL J, MCDONALD W H, et al. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex[J]. Molecular and Cellar Biology, 2006, 26(5): 1617-1630. DOI:10.1128/MCB.26.5.1617-1630.2006 |
| [9] |
POTTS P R. The yin and yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination[J]. DNA Repair, 2009, 8(4): 499-506. DOI:10.1016/j.dnarep.2009.01.009 |
| [10] |
彭莉, 张飞雄. SMC蛋白的结构和功能[J]. 遗传, 2001, 23(2): 173-176. DOI:10.3321/j.issn:0253-9772.2001.02.020 |
| [11] |
KANNO T, BERTA D G, SJOGREN C, et al. The Smc5/6 complex is an ATP-dependent intermolecular DNA linker[J]. Cell Rep, 2015, 12(9): 1471-82. DOI:10.1016/j.celrep.2015.07.048 |
| [12] |
PEBERNARD S, PERRY J J, TAINER J A, et al. Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability[J]. Molecular Biology of the Cell, 2008, 19(10): 4099-4109. DOI:10.1091/mbc.e08-02-0226 |
| [13] |
PRAHASH S, PRAHASH L. Increased spontaneous mitotic segregation in MMS-sensitive mutants of Saccharomyces cerevisiae[J]. Genetics, 1977, 87(2): 229-236. DOI:10.1093/genetics/87.2.229 |
| [14] |
POTTS P R, YU H T. Human MMS21/NSE2 is a SUMO ligase required for DNA repair[J]. Molecular & Cellular Biology, 2005, 25(16): 7021-7032. |
| [15] |
ZHAO X L, BLOBEL G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization[J]. Proceedings of the National Academy of Sciences of the United States of America, 2005, 102(13): 4777-4782. DOI:10.1073/pnas.0500537102 |
| [16] |
TAYLOR E M, COPSEY A C, HUDSON J J R, et al. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex[J]. Molecular and Cellular Biology, 2008, 28(4): 1197-1206. DOI:10.1128/MCB.00767-07 |
| [17] |
DOYLE J M, GAO J L, WANG J W, et al. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases[J]. Molecular Cell, 2010, 39(6): 963-974. DOI:10.1016/j.molcel.2010.08.029 |
| [18] |
CHOMEZ P, BACKER O D, BERTRAND M, et al. An overview of the MAGE gene family with the identification of all human members of the family[J]. Cancer Research, 2001, 61(14): 5544-5551. |
| [19] |
PALECEK J, VIDOT S, FENG M, et al. The Smc5-Smc6 DNA repair complex.Bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits[J]. Journal of Biological Chemistry, 2006, 281(48): 36952-36959. DOI:10.1074/jbc.M608004200 |
| [20] |
DUAN X Y, YANG Y, CHEN Y H, et al. Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5-6 subcomplex and the hinge regions of Smc5 and Smc6[J]. Journal of Biological Chemistry, 2009, 284(13): 8507-8515. DOI:10.1074/jbc.M809139200 |
| [21] |
SERGEANT J, TAYLOR E, PALECEK J, et al. Composition and architecture of the Schizosaccharomyces pombe Rad18(Smc5-6) complex[J]. Molecular and Cellular Biology, 2005, 25(1): 172-184. DOI:10.1128/MCB.25.1.172-184.2005 |
| [22] |
XU P L, YUAN D K, LIU M, et al. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots[J]. Plant Physiology, 2013, 161(4): 1755-1768. DOI:10.1104/pp.112.208942 |
| [23] |
ZABRADY K, ADAMUS M, VONDROVA L, et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA[J]. Nucleic Acids Research, 2016, 44(3): 1064-1079. DOI:10.1093/nar/gkv1021 |
| [24] |
HUDSON J J R, BEDNAROVA K, KOZAKOVA L, et al. Interactions between the Nse3 and Nse4 components of the SMC5-6 complex identify evolutionarily conserved interactions between MAGE and EID Families[J]. PLoS One, 2011, 6(2): e17270. DOI:10.1371/journal.pone.0017270 |
| [25] |
YUAN S P, WANG W, MARQUES J, et al. Salicylic acid activates DNA damage responses to potentiate plant immunity[J]. Molecular Cell, 2013, 52(4): 602-610. DOI:10.1016/j.molcel.2013.09.019 |
| [26] |
TASCHNER M, BASQUIN J, STEIGENBERGER B, et al. Nse5/6 inhibits the Smc5/6 ATPase to facilitate DNA substrate selection[J]. The EMBO Journal, 2021, 40(15): e107807. DOI:10.1101/2021.02.09.430422 |
| [27] |
JEPPSSON K, CARLBORG K K, NAKATO R, et al. The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement[J]. PLoS Genetics, 2014, 10(10): e1004680. DOI:10.1371/journal.pgen.1004680 |
| [28] |
MARIA S R S, GANGAVARAPU V, JOHNSON R E, et al. Requirement of Nse1, a subunit of the Smc5-Smc6 complex, for Rad52-dependent postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae[J]. Molecular and Cellular Biology, 2007, 27(23): 8409-8418. DOI:10.1128/MCB.01543-07 |
| [29] |
MIYABE I, MORISHITA T, HISHIDA T, et al. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe Rad60 and smc5/6 mutants after release from replication arrest[J]. Molecular and Cellular Biology, 2006, 26(1): 343-353. DOI:10.1128/MCB.26.1.343-353.2006 |
| [30] |
PICCOLI G D, CORTES-LEDESMA F, IRA G, et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination[J]. Nature Cell Biology, 2006, 8(9): 1032-1034. DOI:10.1038/ncb1466 |
| [31] |
LINDROOS H B, STROM L, ITOH T, et al. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways[J]. Molecular Cell, 2006, 22(6): 755-767. DOI:10.1016/j.molcel.2006.05.014 |
| [32] |
BERMUDEZ-LOPEZ M, ARAGON L. Smc5/6 complex regulates Sgs1 recombination functions[J]. Current Genetics, 2017, 63(3): 381-388. DOI:10.1007/s00294-016-0648-5 |
| [33] |
BONNER J N, CHOI K, XUE X Y, et al. Smc5/6 Mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates[J]. Cell Reports, 2016, 16(2): 368-378. DOI:10.1016/j.celrep.2016.06.015 |
| [34] |
POTTS P R, PORTEUS M H, YU H T. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks[J]. The EMBO Journal, 2006, 25(14): 3377-3388. DOI:10.1038/sj.emboj.7601218 |
| [35] |
WATANABE K, PACHER M, DUKOWIC S, et al. The structural maintenance of chromosomes 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana[J]. The Plant Cell, 2009, 21(9): 2688-2699. DOI:10.1105/tpc.108.060525 |
| [36] |
STEPHAN A, KLISZCZAK M, DODSON H, et al. Roles of vertebrate Smc5 in sister chromatid cohesion and homologous recombinational repair[J]. Molecular and Cellular Biology, 2011, 31(7): 1369-1381. DOI:10.1128/MCB.00786-10 |
| [37] |
MENGISTE T, REVENKOVA E, BECHTOLD N, et al. An SMC-like protein is required for efficient homologous recombination in Arabidopsis[J]. The EMBO Journal, 1999, 18(16): 4505-4512. DOI:10.1093/emboj/18.16.4505 |
| [38] |
ZAPATKA M, POCINO-MERINO I, HELUANI-GAHETE H, et al. Sumoylation of Smc5 promotes error-free bypass at damaged replication forks[J]. Cell Reports, 2019, 29(10): 3160-3172. DOI:10.1016/j.celrep.2019.10.123 |
| [39] |
VAREJAO N, IBARS E, LASCORZ J, et al. DNA activates the Nse2/Mms21 SUMO E3 ligase in the Smc5/6 complex[J]. The EMBO Journal, 2018, 37(12): e98306. DOI:10.15252/embj.201798306 |
| [40] |
RASCHLE M, SMEENK G, HANSEN R K, et al. DNA repair.Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links[J]. Science, 2015, 348(6234): 1253671. DOI:10.1126/science.1253671 |
| [41] |
LI G, ZOU W X, JIAN L F, et al. Non-SMC elements 1 and 3 are required for early embryo and seedling development in Arabidopsis[J]. Journal of Experimental Botany, 2017, 68(5): 1039-1054. DOI:10.1093/jxb/erx016 |
| [42] |
ZELKOWSKI M, ZELKOWSKA K, CONRAD U, et al. Arabidopsis NSE4 proteins act in somatic nuclei and meiosis to ensure plant viability and fertility[J]. Frontiers in Plant Science, 2019, 10: 774. DOI:10.3389/fpls.2019.00774 |
| [43] |
DIAZ M, PECINKOVA P, NOWICKA A, et al. The SMC5/6 complex subunit NSE4A is involved in DNA damage repair and seed development[J]. The Plant Cell, 2019, 31(7): 1579-1597. DOI:10.1105/tpc.18.00043 |
| [44] |
DINARDO S, VOELKEL K, STERNGLANZ R. DNA topoisomerase Ⅱ mutant of Saccharomyces cerevisiae: Topoisomerase Ⅱ is required for segregation of daughter molecules at the termination of DNA replication[J]. Proceedings of the National Academy of Sciences of the United States of America, 1984, 81(9): 2616-2620. DOI:10.1073/pnas.81.9.2616 |
| [45] |
KEGEL A, BETTS-LINDROOS H, KANNO T, et al. Chromosome length influences replication-induced topological stress[J]. Nature, 2011, 471(7338): 392-396. DOI:10.1038/nature09791 |
| [46] |
BERMEJO R, DOKSANI Y, CAPRA T, et al. Top1and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation[J]. Genes & Development, 2007, 21(15): 1921-36. |
| [47] |
GALEGO-PAEZ L M, TANAKA H, BANDO M, et al. Smc5/6-mediated regulation of replication progression contributes to chromosome assembly during mitosis in human cells[J]. Molecular Biology of the Cell, 2014, 25(2): 302-317. DOI:10.1091/mbc.e13-01-0020 |
| [48] |
MENOLFI D, DELAMARRE A, LENDRONNE A, et al. Essential roles of the Smc5/6 complex in replication through natural pausing sites and endogenous DNA damage tolerance[J]. Molecular Cell, 2015, 60(6): 835-846. DOI:10.1016/j.molcel.2015.10.023 |
| [49] |
WEHRKAMP-RICHTER S, HYPPA R W, PRUDDEN J, et al. Meiotic DNA joint molecule resolution depends on Nse5-Nse6 of the Smc5-Smc6 holocomplex[J]. Nucleic Acids Research, 2012, 40(19): 9633-9646. DOI:10.1093/nar/gks713 |
| [50] |
BUSTARD D E, MENOLFI D, JEPPSSON K, et al. During replication stress, non-SMC element 5(NSE5) is required for Smc5/6 protein complex functionality at stalled forks[J]. Journal of Biological Chemistry, 2012, 287(14): 11374-11383. DOI:10.1074/jbc.M111.336263 |
| [51] |
ORAVCOVA M, GADALETA M C, NIE M, et al. Brc1 promotes the focal accumulation and SUMO ligase activity of Smc5-Smc6 during replication stress[J]. Molecular and Cellular Biology, 2019, 39(2): e00271-18. DOI:10.1128/MCB.00271-18 |
| [52] |
CHAVEZ A, GEORGE V, AGRAWAL V, et al. Sumoylation and the structural maintenance of chromosomes (Smc) 5/6 complex slow senescence through recombination intermediate resolution[J]. Journal of Biological Chemistry, 2010, 285(16): 11922-11930. DOI:10.1074/jbc.M109.041277 |
| [53] |
NOEL J F, WELLINGER R J. Abrupt telomere losses and reduced end-resection can explain accelerated senescence of Smc5/6 mutants lacking telomerase[J]. DNA Repair (Amst), 2011, 10(3): 271-282. DOI:10.1016/j.dnarep.2010.11.010 |
| [54] |
WAN Y K, ZUO X, ZHUO Y, et al. The functional role of SUMO E3 ligase Mms21p in the maintenance of subtelomeric silencing in budding yeast[J]. Biochemical and Biophysical Research Communications, 2013, 438(4): 746-752. DOI:10.1016/j.bbrc.2013.07.096 |
| [55] |
TORRES-ROSELL J, MACHIN F, FARMER S, et al. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions[J]. Nature Cell Biology, 2005, 7(4): 412-9. DOI:10.1038/ncb1239 |
| [56] |
MORADI-FARD S, SARTHI J, TITTEL-ELMER M, et al. Smc5/6 is a telomere-associated complex that regulates Sir4 binding and TPE[J]. PLoS Genetics, 2016, 12(8): e1006268. DOI:10.1371/journal.pgen.1006268 |
| [57] |
HUANG L X, YANG S G, ZHANG S C, et al. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root[J]. The Plant Journal, 2009, 60(4): 666-678. DOI:10.1111/j.1365-313X.2009.03992.x |
| [58] |
ZOU W X, LI G, JIAN L F, et al. The Arabidopsis SMC6A and SMC6B have redundant function in seed and gametophyte development[J]. Journal of Experimental Botany, 2021, 72(13): 4871-4887. DOI:10.1093/jxb/erab181 |
| [59] |
ZHU L F, FERNANDEZ-JIMENEZ N, SZYMANSKA-LEJMAN M, et al. Natural variation identifies SNI1, the SMC5/6 component, as a modifier of meiotic crossover in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America, 2021, 118(33): e2021970118. DOI:10.1073/PNAS.2021970118 |
| [60] |
KIM D H, HARRIS B, WANG F, et al. Mms21 SUMO ligase activity promotes nucleolar function in Saccharomyces cerevisiae[J]. Genetics, 2016, 204(2): 645-658. DOI:10.1534/genetics.115.181750 |
| [61] |
ZHANG J Y, AUGUSTINE R C, SUZUKI M, et al. The SUMO ligase MMS21 profoundly influences maize development through its impact on genome activity and stability[J]. PLoS Genetics, 2021, 17(10): e1009830. DOI:10.1371/JOURNAL.PGEN.1009830 |
| [62] |
TANIURA H, TANABE N, BANDO Y, et al. Nse1 and Nse4, subunits of the Smc5-Smc6 complex, are involved in Dictyostelium development upon starvation[J]. Development, Growth & Differentiation, 2015, 57(6): 430-443. |
| [63] |
DECORSIERE A, MUELLER H, BREUGEL P C, et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor[J]. Nature, 2016, 531(7594): 386-389. DOI:10.1038/nature17170 |
| [64] |
GIBSON R T, ANDROPHY E J. The SMC5/6 complex represses the replicative program of high-risk human papillomavirus Type 31[J]. Pathogens, 2020, 9(10): 786. DOI:10.3390/pathogens9100786 |



